DigiCortex - Biological Neural Network Simulator
Introduction
DigiCortex is a small simulator of the biological neuron networks, which started as a hobby and which is implementing Eugene Izhikevich’s and Brette-Gerstner (AdEx) phenomenological models of spiking neurons, AMPA, NMDA and GABA synaptic receptor kinetics as well as long-term and short-term synaptic plasticity. DigiCortex is using Diffusion Spectrum Imaging (DSI) data from the Human Connectome Project (HCP) for guiding of pyramidal neuron axons through the white matter. Neurons have multiple compartments and axonal bifurcations (where applicable / configurable by the user).
DigiCortex is currently in a very early phase of development so expect lots of updates in the near future.
It is possible to tweak individual parameters of the simulation such as number of neurons, modify the built-in neuron types (fully open, XML-based format of the neuron library), connectivity statistics (neuron synapse formation affinities, percentages of each post-synaptic neuron type) as well as various additional neuronal parameters (e.g. axonal action potential propagation velocities). Neurons will organize into firing patterns corresponding to Delta, Alpha, Beta and Gamma waves. Long term synaptic plasticity (based on STDP) will slowly update the synaptic strengths and this will influence the firing patterns and frequencies of firing.
Please note that DigiCortex is very resource intensive - especially in terms of memory bandwidth (for example: even mild bumps in DDR3 speed would significantly speed-up the simulation). Current speed limits are mostly due to the memory I/O (e.g. ~1.2x real-time for a simulation of 32768 multi-compartment neurons and 1.8 million synapses on the modern dual Intel Xeon E5 2687W with DDR3 RAM operating @2133 MHz) . I plan to implement support for heterogeneous architectures (CPU + GPU) in the near future.
Starting from version 0.95, DigiCortex also contains the support for acceleration on the NVIDIA CUDA-capable GPUs. CUDA optimizations are implemented by Ana Balevic. Ana is currently completing her PhD at Leiden University in Netherlands. Please note that the CUDA compute plug-in is still in the very early stage, not fully optimized and with limited support for DigiCortex features (no STDP, no current injection, no simulated EEG / fMRI BOLD). These missing features will be added in the near future. Despite the early stage of development, CUDA computation running on GK110 is already more than 7 times faster than 16 Xeon E5 CPU cores.
Download the latest version of DigiCortex (no warranty, use it at your own risk!)
Currently, DigiCortex implements following features:
|
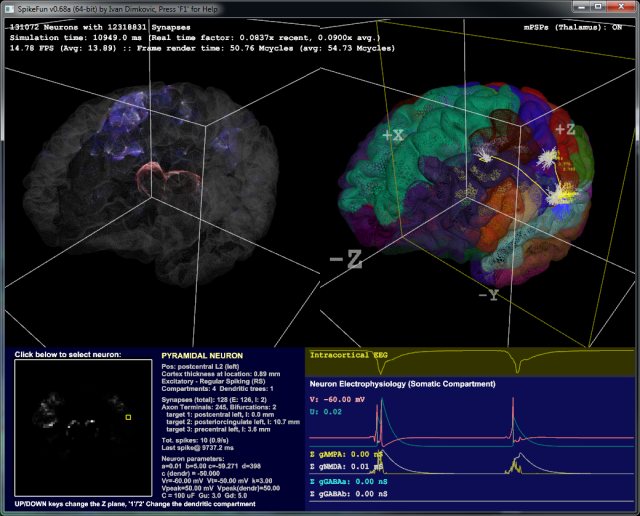
Following picture shows DigiCortex v0.68 in operation ("BrainView"):
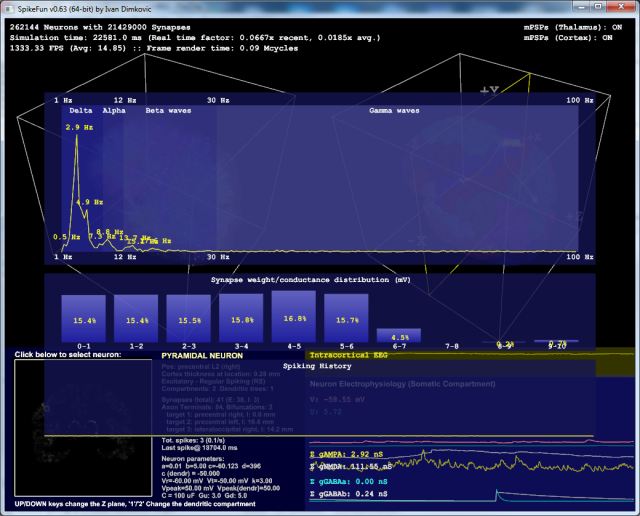
Following picture shows the FFT analysis of the spiking data, simulation of 262144 neurons (press F5 in the simulation for FFT analysis):
Following video shows the visualization-only view with ~1.4 million neurons and ~140 million synapses ('F11' key):
