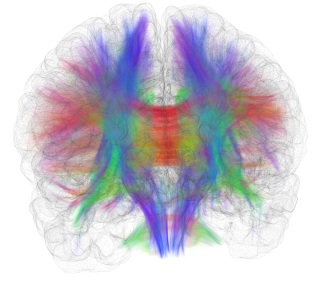
SpikeFun 0.72 - Improved Connectome Mapping
SpikeFun v0.72 contains significantly improved connectome-mapping code, which is greatly improved in precision compared to the earlier versions of SpikeFun.
First of all, the DSI tract tracking library (DigiCortexTractoLib*.dll) has been updated with the latest DSI Studio code base (dated June 4th). Older versions of SpikeFun were using more than a year old code. New code base contains improved tracking algorithms and also adds support for GQI (Generalized Q-Sampling Imaging).
Furthermore the fiber source DSI data is improved, with the externally created white matter mask (through FreeSurfer) and better DSI-T1 data co-registration. Finally, the tract to brain surface mapping algorithm in SpikeFun has been significantly improved in terms of accuracy. Mapping algorithm is now using 12-point affine transformation matrix found during co-registration step and better surface-searching strategies that result in better fitting of tracts to the brain surface.
In under to understand best what is going on, I will try to explain how SpikeFun connectome mapping works on high level. Below diagram shows the workflow that is used in SpikeFun to generate connectome data out of raw structural and diffusion MRI data:
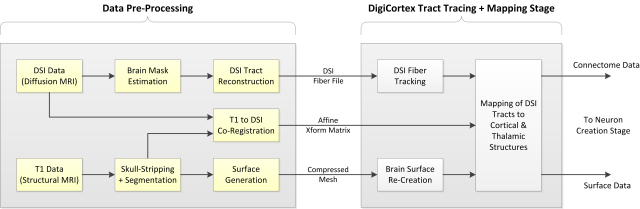
The entire connectome mapping process is divided in two steps – first step is MRI data pre-processing and it is out before SpikeFun is released. This step has two “paths”:
- Structural MRI path (Using T1 MRI data + T2 MRI data to help co-registration)
- Diffusion MRI path (using DSI data)
Structural MRI path is used for brain region segmentation and surface generation. These steps are performed with FreeSurfer. Final surfaces, cortical thickness and segmentation data are stored in appropriate files for usage within SpikeFun. Surface data is compressed so it does not consume large space in the distribution.
DSI path is used for identification of the white matter axonal pathways. In order to avoid artifacts, it is necessary to identify brain vs. non-brain tissue in the diffusion MRI scan. For this, brain mask is created using FreeSurfer. DSI data along with the brain mask is imported in DSI Studio, which performs DSI tract reconstruction (click here for more details on the actual process).
At this stage, T1 and DSI (B0) volumes are co-registered so that structural and connectivity data is properly aligned and free of distortions. The result of this step is a 12-point affine transformation matrix, which is used by SpikeFun during the connectome mapping stage.
At the end of the first stage, data is stored and included in SpikeFun distribution.
Second stage begins when SpikeFun simulation is started. DSI fibers are tracked using DSI Studio tract tracking implementation and brain surface is re-created. Both data streams (tracts and surface) are used by the connectome mapping stage.
Connectome mapping stage transforms the data so tracts and surface are perfectly aligned and then executes the tract-to-surface mapping. Each DSI tract is assigned with the source and destination region, closest triangles in the mesh etc. This data is used to compute region connectivity statistics that will be used during later stages of simulation (neuronal network creation). Tract data is also stored as it will be used in visualization.
Connectome mapping code is heavily multi-threaded, it is able to spawn up to 256 threads (1 thread per logical CPU) and it will reliably pin all the CPUs to 100% during the mapping process. To help with running simulations on slower systems, there is a command line option (-dsimaplevel) which can be used to relax the search algorithm. Of course, reducing number of fibers to track also help on low-end systems, however if not enough fibers are generated it can happen that parts of brain regions do not connect to any other regions due to lack of any guide tracts.
- Log in to post comments